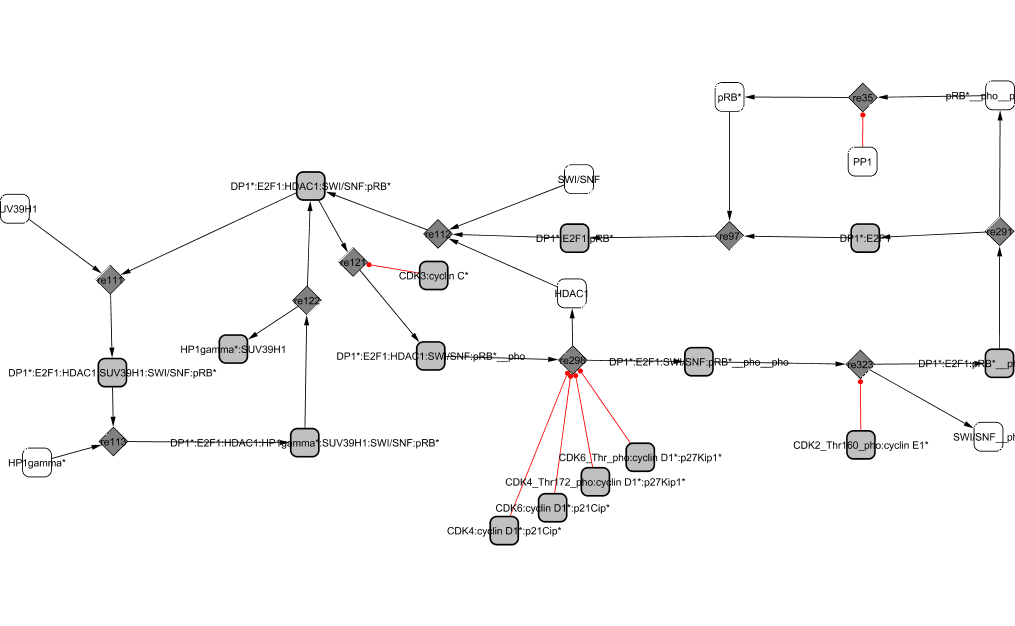
Two cycles appear in this module revealing two roles of RB as (1) a transcriptional repressor: HDAC1 is a marker of transcriptional repression and seems to require the chromatin remodeler SWI-SNF to form with DP1/E2F1/RB a repressor complex (Frolov and Dyson, 2004). The complex recruits both HP1-gamma and SUV39H1 and continues to prevent transcription (Nielsen et al., 2001); (2) a repressor of E2F1/DP1: RB binds to E2F1/DP1 and blocks its transcriptional activity. The affinity between E2F1/DP1 and RB is decreased through sequential phosphorylations by the Cyclin/CDK complexes. The first phosphorylations by CycC/CDK3 favours the passage from G0 to G1 phase, then CycD1/CDK4,6 modifies its conformation and frees HDAC1 (Zhang et al., 2000) revealing a new site of phosphorylation targeted by CycE1/CDK2 (Muchardt and Yaniv, 2001; Vidal and Koff, 2000). The phosphorylation by CycD1/CDK4,6 already allows some genes to be transcribed (such as Cyclin E). When RB is hyperphosphorylated, the complex dissociates and E2F1/DP1 is freed from the inactive complex it was forming with RB protein. Later, RB is dephosphorylated by the phosphatase PP1 towards the end of M phase and able to repress E2Fs again (Vidal and Koff, 2000).
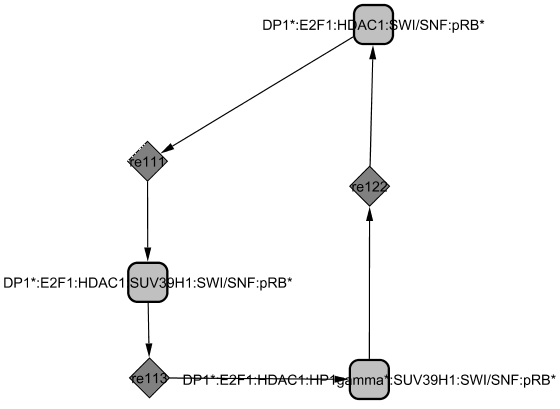 (1)
(1)
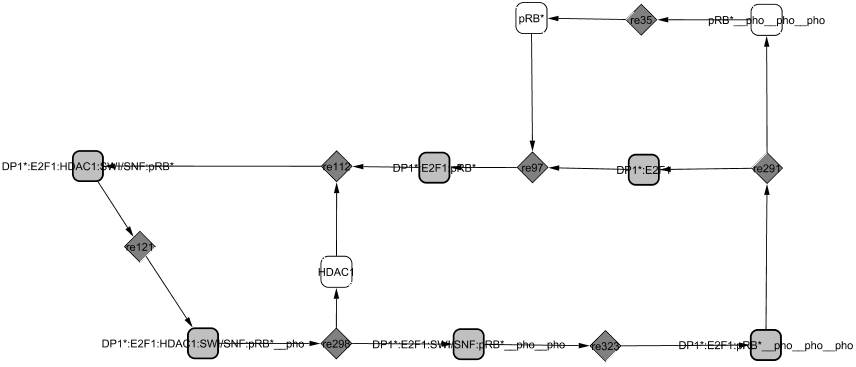 (2)
(2)

