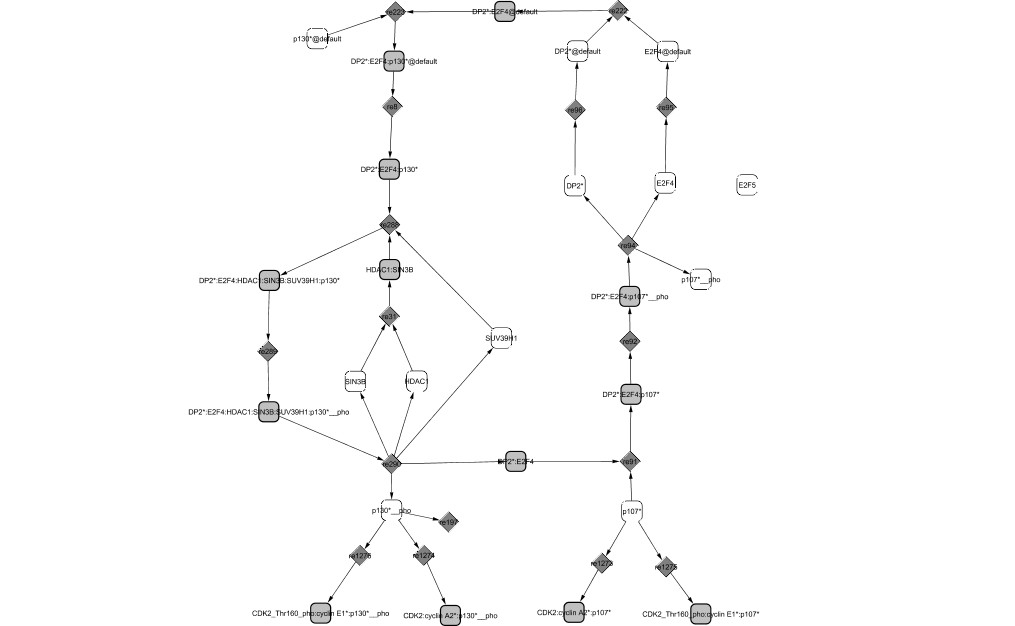
E2F4 associates successively with two different pocket proteins: p130 in quiescent cells and p107 in proliferating cells. E2F4 is initially found in the cytoplasm. When in complex, E2F4 is translocated in the nucleus where it acts as a repressor of transcription during G0 and G1 phases (Verona et al., 1997). Once in the nucleus, the complex binds to DNA and recruits some co-repressors of transcription: the chromatin remodeler Sin3B, the deacetylase histone HDAC1 and the methyltransferase histones SUV39H1 (Liu et al., 2005; Rayman et al., 2002).
At the G0/G1 transition, when the quiescent cells receive signals from growth factors, p130 starts to be phosphorylated and gets dissociated from the complex it was forming with E2F4/DP2. Later, p130 can also be phosphorylated by CycD1/CDK4,6 and CycE1/CDK2 when present and degraded in late G1. It is then replaced by p107 (Farkas et al., 2002). When E2F4/DP2 is in complex with p107, it continues to repress transcription of target genes until it is phosphorylated by CycD1/CDK4,6. E2F4 is then translocated to the cytoplasm where it can no longer repress transcription whereas both p130 and p107 are able to inhibit CycE1/CDK2 and CycA2/CDK2 activities (Litovchick et al., 2004).
In this module, E2F3b and E2F5 should also be considered. Their repressive role in the pathway is not described yet but will be detailed in future versions of the pathway

