The E2F1 module includes the E2F activator family of transcription E2F1, E2F2 and E2F3a. Even though the proteins slightly differ in their cell cycle role, we chose to describe the three E2F transcription factors as one since they share a lot of functional similarities. In a future version of the pathway, we plan to differentiate the activity of the three transcription activators.
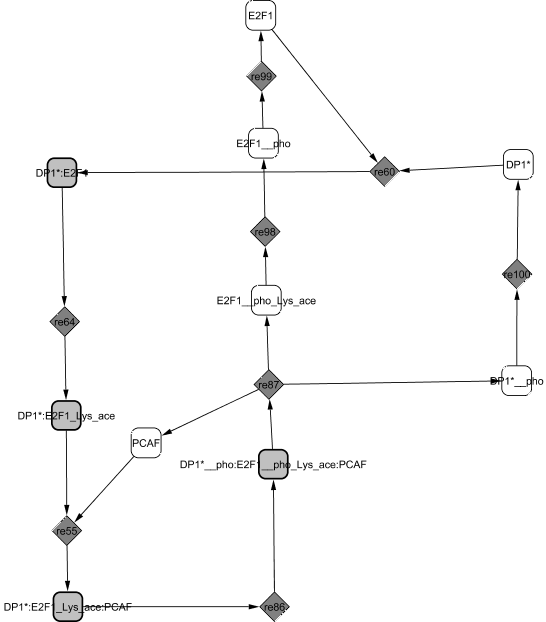
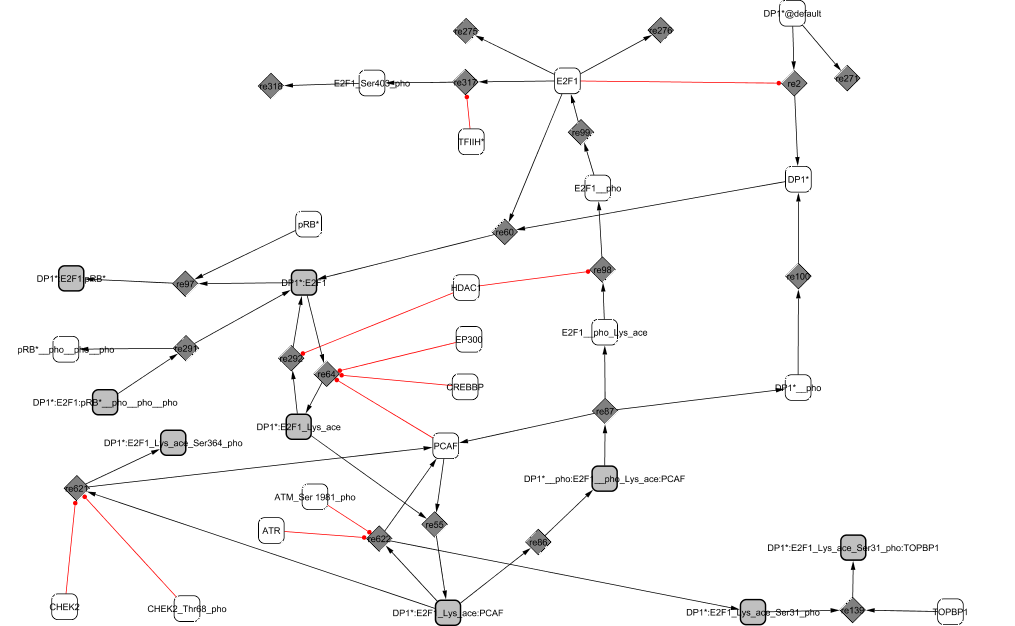
Experiments show that dimerization between E2F1 and its partner DP1 is stable and that E2F1 stimulates nuclear localization of DP1 (Magae et al., 1996). E2F1/DP1 is acetylated by the three acetyltransferases PCAF, CREB Binding Protein and p300 which stabilizes E2F1 protein (Frolov and Dyson, 2004). The acetylated complex is able to bind to PCAF to form an active dimer. The complex ability to bind to DNA on the promoter sites of its target genes along with its transcriptional activity are increased at the G1/S transition.
During G2, the complex is phosphorylated by CycA2/CDK2 (He and Cress, 2002). The affinity between E2F1 and DP1 is then diminished leading to the dissociation of the complex (Tsantoulis and Gorgoulis, 2005) and the release of PCAF. The proteins undergo further modifications before degradation: E2F1 is deactelylated by HDAC1 (Martinez-Balbas et al., 2000), dephosphorylated and phosphorylated de novo during S phase by TFIIH kinase for rapid degradation (Ianari et al., 2004).
Upon DNA damage, the complex PCAF/E2F1/DP1 can be phosphorylated and stabilized either by CHEK1 and CHEK2 through phosphorylation at Ser-364, or by ATM and ATR (Dimova and Dyson, 2005; Powers et al., 2004), preventing E2F1 ubiquitination (Wang et al., 2004). E2F1 mediates the transcription of many genes involved in apoptosis. However, E2F1 transcriptional activity can also be inhibited when bound to the topoisomerase TopBP1 in order to give time to the cell to repair the damage (Liu et al., 2003).