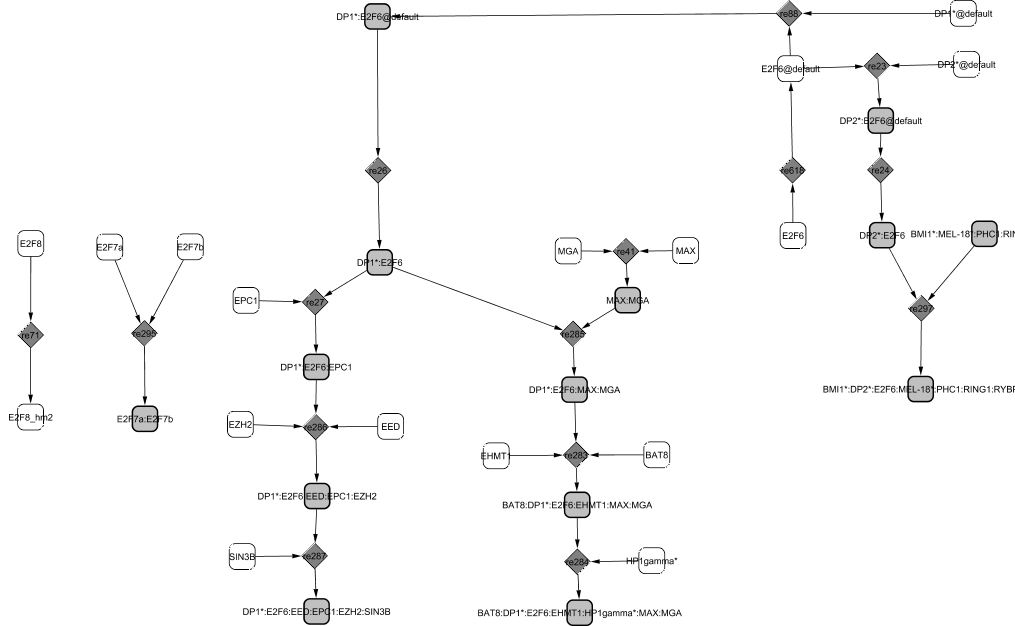
E2F6 seems to play a role in S phase entry. It binds to both DP partners, DP1 and DP2 in the cytoplasm and the complexes are then translocated in the nucleus. As opposed to other E2F family members, E2F6 does not associate with pocket proteins but rather recruits some proteins from the polycomb group (PcG) to repress transcription.
The first polycomb group with which E2F6 is involved includes BMI1, Mel-18, PHC1, RING1, and RYBP and acts as a repressor of transcription (Sánchez-Beato et al., 2004; Trimarchi et al., 2001). Similarly, E2F6, Max and HP1-gamma bind to form a complex that has also revealed a repressive transcription role in quiescent cells (Ogawa et al., 2002). However, E2F6 seems to intervene in other transitions of the cell cycle than only that of G0/G1. Another complex involving EPC1 and EZH2 has been found in proliferating cells: E2F6/DP1 is repressing transcription when in complex with EPC1 alone or with both EPC1 and EZH2 (Attwooll et al., 2005). E2F6/DP1 binds consecutively to EPC1 and then to both EZH2 and EED. The roles of the different complexes are not clearly established yet and more experiments will be needed to describe confidently their specific actions in proliferating and quiescent cells.
E2F7 and E2F8 regulations have not been carefully defined in our pathway yet. Both are known to inhibit transcription though (de Bruin et al., 2003; Logan et al., 2005). The details will be added as more publications on their role in RB pathway appear. |

