

Next: 3.3.4 Clustering profiles
Up: 3.3 Data analysis
Previous: 3.3.2 FrAGL (Frequency of
Contents
3.3.3 Finding common alterations among a collection of CGH- array profiles
Instead of looking for individual probes carrying genome alterations, it is often fruitful to consider the geography of the genome and to look for whole regions. Rouveirol et al. (2006) have described algorithms for finding common alterations. Two algorithms are proposed within VAMP. It is necessary that breakpoints, gain, loss and amplicon regions have been previously detected by any algorithm (Hupé et al., 2004). We propose two different approaches to identify such regions of biological interest:
- Tools
 Genome Alteration
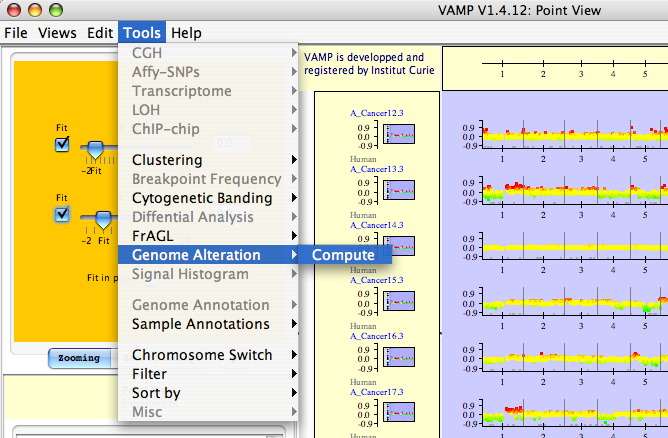
Genome Alteration  Compute: Minimal Alterations are extracted by intersecting the profiles of many tumors and looking for a sufficient number of alterations in the tumors (this parameter is set by the user) over the smallest possible region of the profile (see Figures 3.37, 3.38 and 3.39).
Compute: Minimal Alterations are extracted by intersecting the profiles of many tumors and looking for a sufficient number of alterations in the tumors (this parameter is set by the user) over the smallest possible region of the profile (see Figures 3.37, 3.38 and 3.39).
Figure 3.37:
Tools  Genome Alteration
Genome Alteration  Compute - The user opens a new dialog window to set the parameters for Minimal Alterations.
Compute - The user opens a new dialog window to set the parameters for Minimal Alterations.
|
|
Figure 3.38:
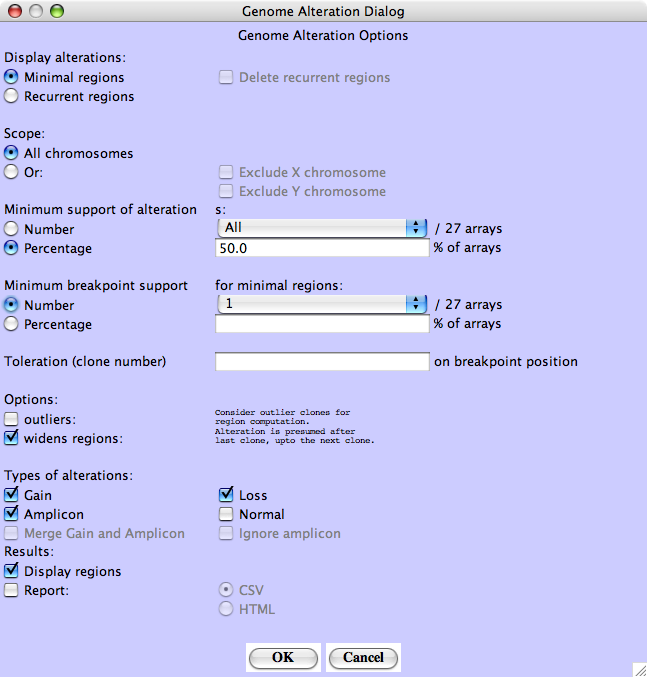
Minimal Alterations - Choose Minimal regions - At least 50% of the profiles must share the same alterations.
|
|
Figure 3.39:
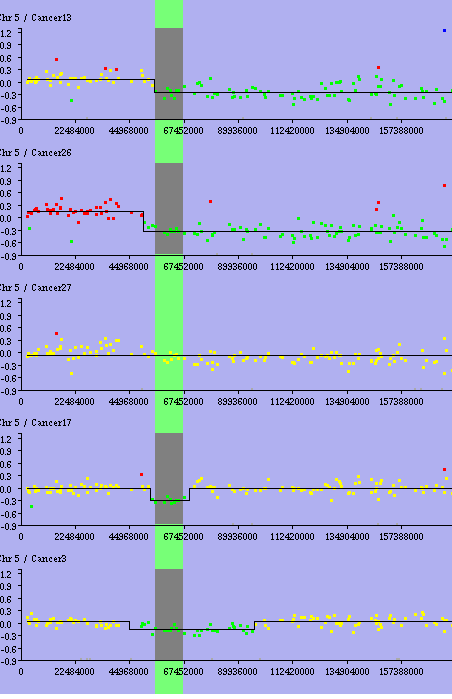
Minimal Alterations - Results : the minimal alterations are drawn in red for gain, green for loss. Here there is only one loss region.
|
|
- Tools
 Genome Alteration
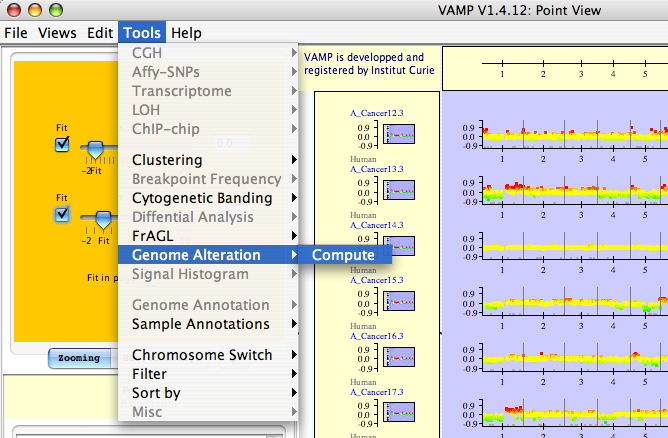
Genome Alteration  Compute: In a given tumor, an alteration is bounded by two extremities, which can be a breakpoint or a chromosome end; when an alteration is present in a sufficient number of tumors with the same extremities, it is a recurrent alteration (see Figures 3.40, 3.41 and 3.42).
Compute: In a given tumor, an alteration is bounded by two extremities, which can be a breakpoint or a chromosome end; when an alteration is present in a sufficient number of tumors with the same extremities, it is a recurrent alteration (see Figures 3.40, 3.41 and 3.42).
Figure 3.40:
Tools  Genome Alteration
Genome Alteration  Compute - The user opens a new dialog window to set the parameters for Recurrent Alterations.
Compute - The user opens a new dialog window to set the parameters for Recurrent Alterations.
|
|
Figure 3.41:
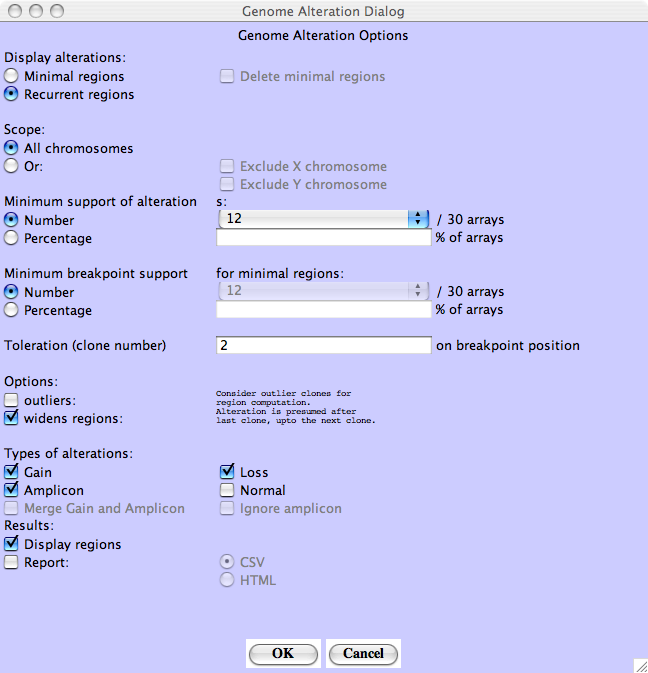
Recurrent Alterations - Choose Recurrent regions - At least 12 out of 30 profiles must share the same alterations.
|
|
Figure 3.42:
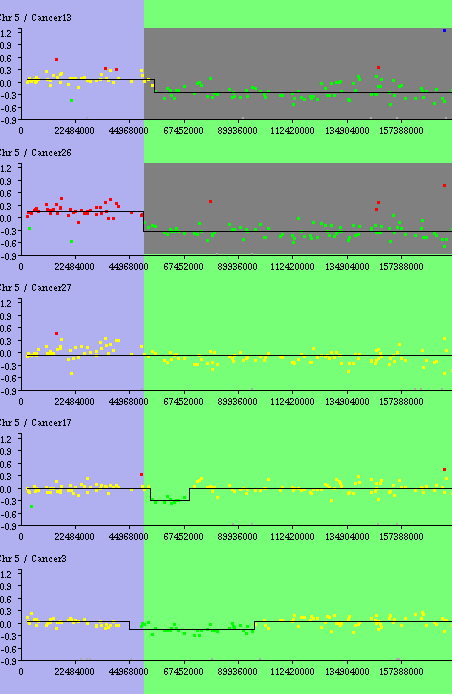
Recurrent Alterations - Results : the recurrent alterations are drawn in red for gain, green for loss. Here there is only one loss region.
|
|
For both Recurrent Alterations and Minimal Alterations, the user has to set the Minimum support required, that is the minimum number or percentage of tumors showing the alteration for considering it as significant.



Next: 3.3.4 Clustering profiles
Up: 3.3 Data analysis
Previous: 3.3.2 FrAGL (Frequency of
Contents
2007 - Institut Curie Bioinformatics unit