RB has been found to associate with many partners (110 cellular proteins), either activating or repressing them. In our pathway, we concentrate on RB and its partners that are directly related to cell cycle events.
According to Morris and Dyson (2001,
Adv Cancer Res. 82:1-54), these proteins can be divided in three groups:
- Phosphatases or kinases that alter RB function: 15 protein kinases or phosphatases affect RB function by phosphorylating or dephosphorylating them.
- Transcriptional regulators that can either repress or activate transcription: 72 transcriptional factors bind to RB. The complexes affect transcription in different ways. A list of the activators or repressors of transcription can be found in Morris & Dyson or by clicking on the diagram below.
- Other types of proteins that seem to be regulated by RB through association but whose funtions are not clear yet: 23 of these factors found to bind to RB have diverse functions in the cell. Among them, can be found proteins related to cell cycle or DNA regulation, heatshock proteins, proteins from the nuclear matrix, etc.
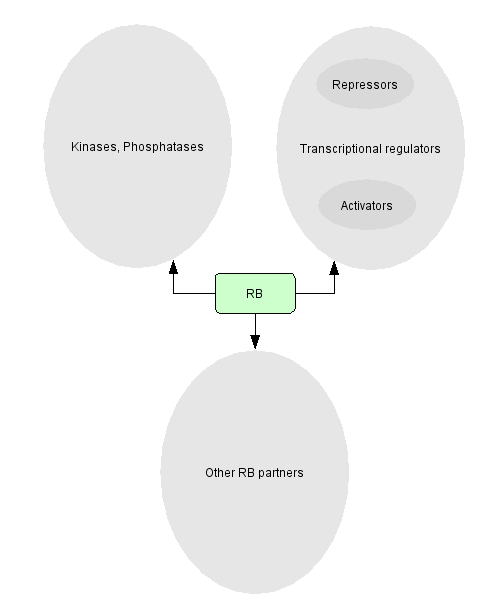
By clicking on each subgroup, the complete lists of factors appear. The ones that are in red are in our description of RB pathway .

