To retrieve the list of genes located in the amplified regions previously identified do the following:
A dialog box appear and set the parameters as follows (see Figure 4.8(c)):
Now you can open a new web browser window and open the HTML file gene-list.html to visualize all the genes located in the amplified regions (see Figure 4.9).
|
[Human gene import] 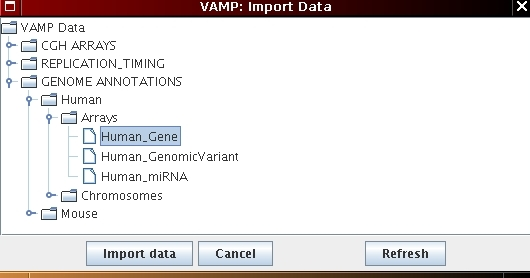 [Tools menu - Genome Annotations]
[Tools menu - Genome Annotations]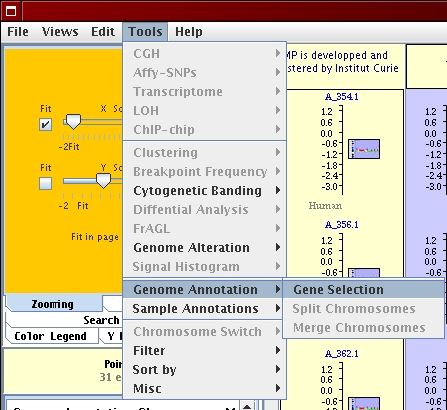 [Genome Annotations parameters]
[Genome Annotations parameters]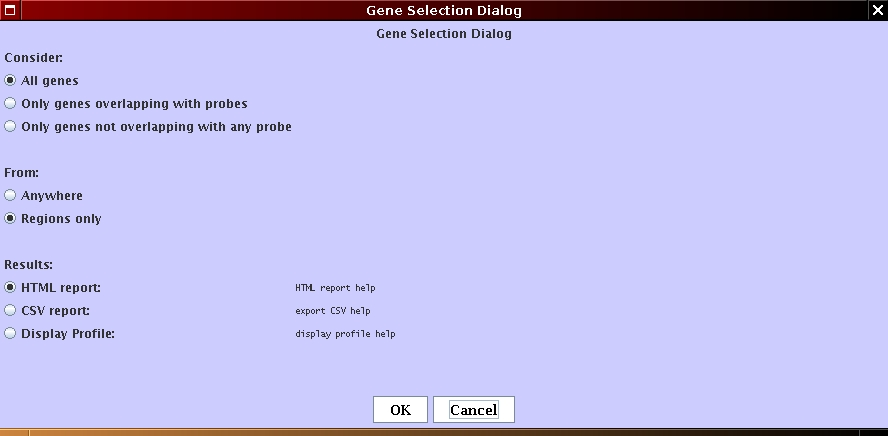
|