First, start the VAMP software (La Rosa et al., 2006) from the web-page:
http://bioinfo.curie.fr/actudb/access.php.
Then, load the array CGH profiles as follows:
n.b.: if you wish to import data from one specific chromosome select:
CGH ARRAYS/douglas_2004/Chromosomes/chr.. (see Figure 5.1(c))
|
[File menu] 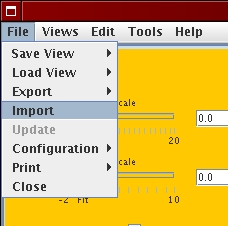 [Genome wide profile import]
[Genome wide profile import]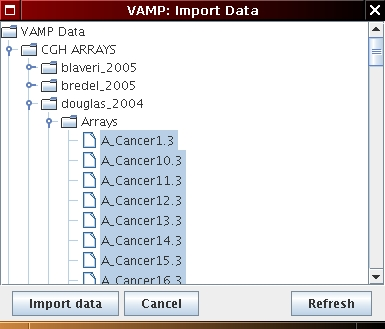 [Chromosome profile import (optional)]
[Chromosome profile import (optional)]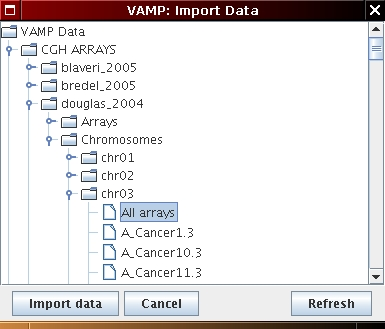
|
The genomic profiles are displayed in the interface as shown in Figure 5.2.
Open a new view (see Figure 5.3) and repeat the previous steps for the nakao_2004 directory.