SHARP Pathway
Database
Institut
Curie node:
Coordinator: Patrick Poullet.
Participants: Amélie Gelay, Andrei Zinovyev, Philippe La Rosa and Emmanuel Barillot.
Collaborators: François Radvanyi, Olivier Delattre, Yohanns Bellaiche and Jean
de-Gunzburg.
SHARP global coordinator: Ron Shamir (School of Computer
Science, Tel Aviv University, Israel).
The SHARP project:
Making sense of the intricate network
of proteins and genes regulations that take place inside the cell during cell
growth, differentiation and apoptosis is a critical step in understanding how
these processes are controlled at the molecular level. Unfortunately, the
biological knowledge collected so far is fragmented in thousands of research
articles of heterogeneous quality making it difficult for researchers to
comprehend globally the mechanisms involved. This knowledge must be centralized
and standardized to become easily accessible. Furthermore, as the data
accumulate, it becomes necessary to use computer power to store, visualize
and analyze these complex regulations.
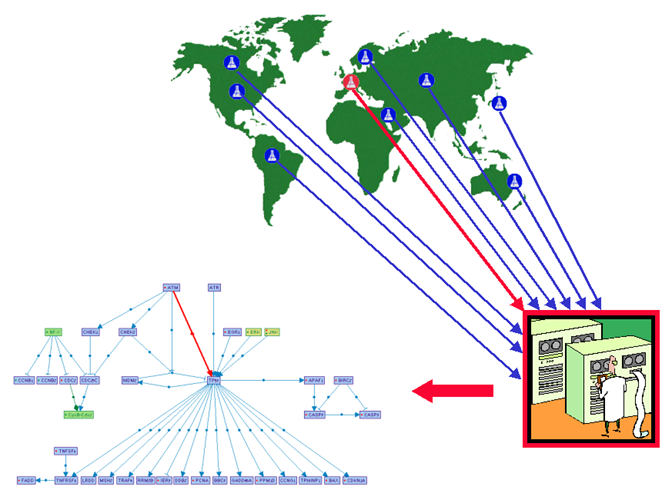
The SHARP consortium was initiated
by Ron Shamir at Tel Aviv University to address this problem using a
collaborative approach. Researchers from all around the world -including from
the Institut Curie- have already joined this consortium. The goal of this
global collaboration is to take advantage of each research group’s expertise on
specific signaling pathways to collect, curate and formalize the data available
from the literature or from their own laboratories. New data are regularly
integrated to a central pathway database and made available to all members of
the consortium (see figure below).
A SHARP software was designed
and developed by Ron Shamir’s group to assist researchers through this task.
This tool consists of a multi-platform (java-based) intuitive pathway
visualization module which communicates with an underlying database. The user
can use this interface to submit new data and to query, edit and analyze the
data stored. This tool can also be used to superimposed external quantitative
data such as gene expression data (microarrays) over the signaling
pathways.
Our task is to provide technical
support to the research groups of the Institut Curie involved in the SHARP
project and to coordinate their work with that of the other members of the
consortium. The data collected are then used in other projects of the Group
in particular for the analysis of microarray data
(e.g. Kernelchip).
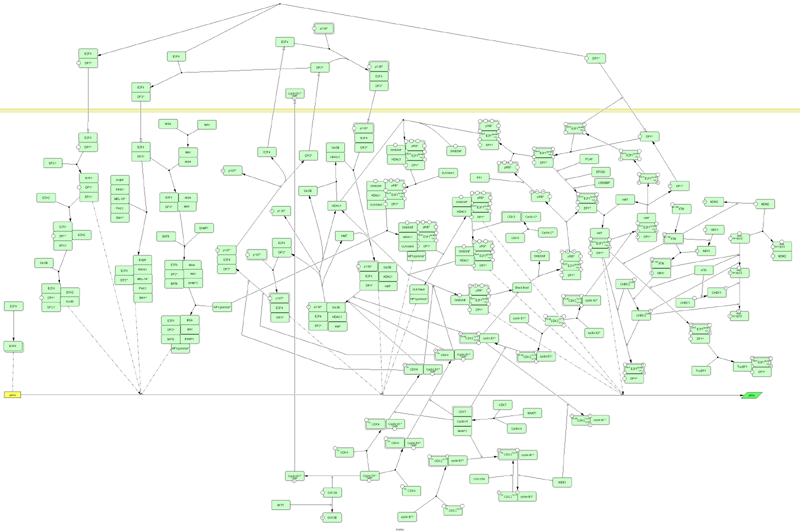
The Retinoblastoma (RB) Pathway case:
Participants: Amélie Gelay, François Radvanyi,
Andrei Zinovyev, Emmanuel Barillot.
The purpose of the present work is
to collect as much information as possible on gene-regulatory networks and
organize it in an exploitable way for the analysis of data from microarrays. We
started with RB, a tumor suppressor gene involved in a pathway controlling the
cell cycle progression, which is one of the most frequent targets of genetic
alterations in human cancer.
The data are collected from public
sources (reviews, publications…) and stored using CellDesigner. CellDesigner is
a structured diagram editor for drawing gene-regulatory and biochemical
networks. These networks are drawn based on the process diagram, with the
graphical notation system proposed by Kitano, and are stored using the Systems
Biology Markup Language (SBML). SBML is a standard computer-readable format for
representing models of biochemical reaction networks. It is applicable to
metabolic networks, cell-signaling pathways, regulatory networks, and many
others. Once converted in SBML format, signaling pathway data can be imported
in network visualization tools such as SHARP.
References for CellDesigner:
1.
Kitano, H., Funahashi, A., Matsuoka, Y., Oda, K. (2005) Using process
diagrams for the graphical representation of biological networks. Nat Biotechnol. 23(8):961-6.
2.
Kitano, H. (2003) A graphical notation for biochemical networks. BIOSILICO. 1. 169-176.
3.
Hucka, M., Finney, A., Sauro, H.M., Bolouri, H., Doyle, J.C., Kitano,
H., Arkin, A.P., Bornstein, B.J., Bray, D., Cornish-Bowden, A., Cuellar, A.A.,
Dronov, S., Gilles, E.D., Ginkel, M., Gor, V., Goryanin, I.I., Hedley, W.J.,
Hodgman, T.C., Hofmeyr, J.H., Hunter, P.J., Juty, N.S., Kasberger, J.L.,
Kremling, A., Kummer, U., Le Novere, N., Loew, L.M., Lucio, D., Mendes, P.,
Minch, E., Mjolsness, E.D., Nakayama, Y., Nelson, M.R., Nielsen, P.F.,
Sakurada, T., Schaff, J.C., Shapiro, B.E., Shimizu, T.S., Spence, H.D.,
Stelling, J., Takahashi, K., Tomita, M., Wagner, J., Wang, J.; SBML Forum.
(2003) The systems biology markup language (SBML): a medium for representation
and exchange of biochemical network models. Bioinformatics.
19(4):524-31
4.
Kitano, H. (2002) Systems biology: a brief overview. Science 295(5560):1662-4.
References for the Retinoblastoma pathway:
1.
Attwooll, C., Lazzerini Denchi, E., Helin, K. (2004) The E2F family:
specific functions and overlapping interests. EMBO J. 23(24):4709-16.
2.
Attwooll, C., Oddi, S., Cartwright, P., Prosperini, E., Agger, K.,
Steensgaard, P., Wagener, C., Sardet, C., Moroni, M.C., Helin, K. (2005) A
novel repressive E2F6 complex containing the polycomb group protein, EPC1, that
interacts with EZH2 in a proliferation-specific manner. J Biol Chem. 280(2):1199-208.
3.
Bartkova, J., Horejsi, Z., Koed, K., Kramer, A., Tort, F., Zieger, K.,
Guldberg, P., Sehested, M., Nesland, J.M., Lukas, C., Orntoft, T., Lukas, J.,
Bartek, J. (2005) DNA damage response as a candidate anti-cancer barrier in
early human tumorigenesis. Nature.
434(7035):864-70.
4.
Blais, A., Dynlacht, B.D. (2004) Hitting their targets: an emerging
picture of E2F and cell cycle control. Curr
Opin Genet Dev. 14(5):527-32.
5.
Boyer Arnold, N., Korc, M. (2005) Smad7 abrogates transforming growth
factor-beta1-mediated growth inhibition in COLO-357 cells through functional
inactivation of the retinoblastoma protein. J
Biol Chem. 280(23):21858-66.
6.
Brehm, A., Miska, E.A., McCance, D.J., Reid, J.L., Bannister, A.J.,
Kouzarides, T. (1998) Retinoblastoma protein recruits histone deacetylase to
repress transcription. Nature.
391(6667):597-601.
7.
Brown, VD., Phillips, R.A., Gallie, B.L. (1999) Cumulative effect of
phosphorylation of pRB on regulation of E2F activity. Mol Cell Biol. 19(5):3246-56.
8.
Cam, H., Dynlacht, B.D. (2003) Emerging roles for E2F: beyond the G1/S
transition and DNA replication. Cancer
Cell. 3(4):311-6.
9.
Chan, H.M., La Thangue, N.B. (2001) p300/CBP proteins: HATs for
transcriptional bridges and scaffolds. J
Cell Sci. 114(Pt 13):2363-73.
10. Claudio, PP.,
Tonini, T., Giordano, A. (2002) The retinoblastoma family: twins or distant
cousins? Genome Biol. 3(9):reviews3012.
11. Cobrinik, D.
(2005) Pocket proteins and cell cycle control. Oncogene. 24(17):2796-809.
12. Coqueret, O.
(2002) Linking cyclins to transcriptional control. Gene. 299(1-2):35-55.
13. Dahiya, A., Wong,
S., Gonzalo, S., Gavin, M., Dean, D.C. (2001) Linking the Rb and polycomb
pathways. Mol Cell. 8(3):557-69.
Erratum in: Mol Cell. 2003
Jun;11(6):1693-6.
14. Deng, CX.,
Brodie, S.G. (2000) Roles of BRCA1 and its interacting proteins. Bioessays. 22(8):728-37
15. Di Stefano, L.,
Jensen, M.R., Helin, K. (2003) E2F7, a novel E2F featuring DP-independent
repression of a subset of E2F-regulated genes. EMBO J. 22(23):6289-98.
16. Dimova, D.K.,
Dyson, N.J. (2005) The E2F transcriptional network: old acquaintances with new
faces. Oncogene. 24(17):2810-26.
17. Dimova, D.K.,
Stevaux, O., Frolov, M.V., Dyson, N.J. (2003) Cell cycle-dependent and cell
cycle-independent control of transcription by the Drosophila E2F/RB pathway. Genes Dev. 17(18):2308-20.
18. DuPree, E.L.,
Mazumder, S., Almasan, A. (2004) Genotoxic stress induces expression of E2F4, leading
to its association with p130 in prostate carcinoma cells. Cancer Res. 64(13):4390-3.
19. Fang, W., Mori,
T., Cobrinik, D. (2002) Regulation of PML-dependent transcriptional repression
by pRB and low penetrance pRB mutants. Oncogene.
21(36):5557-65.
20. Farkas, T.,
Hansen, K., Holm, K., Lukas, J., Bartek, J. (2002) Distinct phosphorylation
events regulate p130- and p107-mediated repression of E2F-4. J Biol Chem. 277(30):26741-52.
21. Ferreira, R.,
Naguibneva, I., Mathieu, M., Ait-Si-Ali, S., Robin, P., Pritchard, L.L.,
Harel-Bellan, A. (2001) Cell cycle-dependent recruitment of HDAC-1 correlates
with deacetylation of histone H4 on an Rb-E2F target promoter. EMBO Rep. 2(9):794-9.
22. Ferreira, R.,
Naguibneva, I., Pritchard, L.L., Ait-Si-Ali, S., Harel-Bellan, A. (2001) The
Rb/chromatin connection and epigenetic control: opinion. Oncogene. 20(24):3128-33.
23. Fortin, A.,
MacLaurin, J.G., Arbour, N., Cregan, S.P., Kushwaha, N., Callaghan, S.M., Park,
D.S., Albert, P.R., Slack, R.S. (2004) The proapoptotic gene SIVA is a direct
transcriptional target for the tumor suppressors p53 and E2F1. J Biol Chem. 279(27):28706-14.
24. Frolov, M.V.,
Dyson, N.J. (2004) Molecular mechanisms of E2F-dependent activation and
pRB-mediated repression. J Cell Sci.
117(Pt 11):2173-81.
25. Gagrica, S., Hauser,
S., Kolfschoten, I., Osterloh, L., Agami, R., Gaubatz, S. (2004) Inhibition of
oncogenic transformation by mammalian Lin-9, a pRB-associated protein. EMBO J. 23(23):4627-38.
26. Ginsberg, D.
(2004) E2F3-a novel repressor of the ARF/p53 pathway. Dev Cell. 6(6):742-3.
27. Grafstrom, RH.,
Pan, W., Hoess, R.H. (1999) Defining the substrate specificity of cdk4
kinase-cyclin D1 complex. Carcinogenesis.
20(2):193-8.
28. Gunawardena,
R.W., Siddiqui, H., Solomon, D.A., Mayhew, C.N., Held, J., Angus, S.P.,
Knudsen, E.S. (2004) Hierarchical requirement of SWI/SNF in retinoblastoma
tumor suppressor-mediated repression of Plk1. J Biol Chem. 279(28):29278-85.
29. Hitchens, M.R.,
Robbins, P.D. (2003) The role of the transcription factor DP in apoptosis. Apoptosis. 8(5):461-8.
30. Ianari, A.,
Gallo, R., Palma, M., Alesse, E., Gulino, A. (2004) Specific role for
p300/CREB-binding protein-associated factor activity in E2F1 stabilization in
response to DNA damage. J Biol Chem.
279(29):30830-5.
31. Ishida, H.,
Masuhiro, Y., Fukushima, A., Argueta, J.G., Yamaguchi, N., Shiota, S.,
Hanazawa, S. (2005) Identification and characterization of novel isoforms of
human DP-1: DP-1{alpha} regulates the transcriptional activity of E2F1 as well
as cell cycle progression in a dominant-negative manner. J Biol Chem. 280(26):24642-8.
32. Jacobs, JJ., van
Lohuizen, M. (2002) Polycomb repression: from cellular memory to cellular
proliferation and cancer. Biochim Biophys
Acta. 1602(2):151-61.
33. Jhanwar-Uniyal,
M. (2003) BRCA1 in cancer, cell cycle and genomic stability. Front Biosci. 8:s1107-17.
34. Johnson, DG.,
Schneider-Broussard, R. (1998) Role of E2F in cell cycle control and cancer. Front Biosci. 3:d447-8
35. Knudsen, ES.,
Wang, J.Y. (1997) Dual mechanisms for the inhibition of E2F binding to RB by
cyclin-dependent kinase-mediated RB phosphorylation. Mol Cell Biol. 17(10):5771-83.
36. Kyrylenko, S.,
Korhonen, P., Kyrylenko, O., Roschier, M., Salminen, A. (2000) Expression of
transcriptional repressor proteins mSin3A and 3B during aging and replicative
senescence. Biochem Biophys Res Commun.
275(2):455-9.
37. Lin, WC., Lin,
F.T., Nevins, J.R. (2001) Selective induction of E2F1 in response to DNA
damage, mediated by ATM-dependent phosphorylation. Genes Dev. 15(14):1833-44.
38. Logan, N.,
Delavaine, L., Graham, A., Reilly, C., Wilson, J., Brummelkamp, T.R., Hijmans,
E.M., Bernards, R., La Thangue, N.B. (2004) E2F-7: a distinctive E2F family
member with an unusual organization of DNA-binding domains. Oncogene. 23(30):5138-50.
39. Logan, N.,
Graham, A., Zhao, X., Fisher, R., Maiti, B., Leone, G., La Thangue, N.B. (2005)
E2F-8: an E2F family member with a similar organization of DNA-binding domains
to E2F-7. Oncogene. 24(31):5000-4.
40. Ludlow, JW.,
Glendening, C.L., Livingston, D.M., DeCarprio, J.A. (1993) Specific enzymatic
dephosphorylation of the retinoblastoma protein. Mol Cell Biol. 13(1):367-72.
41. Lund, A.H., van
Lohuizen, M. (2004) Polycomb complexes and silencing mechanisms. Curr Opin Cell Biol. 16(3):239-46.
42. Magae, J., Wu,
C.L., Illenye, S., Harlow, E., Heintz, N.H. (1996) Nuclear localization of DP
and E2F transcription factors by heterodimeric partners and retinoblastoma
protein family members. J Cell Sci.
109 (Pt7):1717-26.
43. Magnaghi-Jaulin,
L., Groisman, R., Naguibneva, I., Robin, P., Lorain, S., Le Villain, J.P.,
Troalen, F., Trouche, D., Harel-Bellan, A. (1998) Retinoblastoma protein
represses transcription by recruiting a histone deacetylase. Nature. 391(6667):601-5.
44. Maiti, B., Li,
J., de Bruin, A., Gordon, F., Timmers, C., Opavsky, R., Patil, K., Tuttle, J.,
Cleghorn, W., Leone, G. (2005) Cloning and characterization of mouse E2F8, a
novel mammalian E2F family member capable of blocking cellular proliferation. J Biol Chem. 280(18):18211-20.
45. Muchardt, C.,
Yaniv, M. (2001) When the SWI/SNF complex remodels...the cell cycle. Oncogene. 20(24):3067-75.
46. Muller, H.,
Bracken, A.P., Vernell, R., Moroni, M.C., Christians, F., Grassilli, E.,
Prosperini, E., Vigo, E., Oliner, J.D., Helin, K. (2001) E2Fs regulate the
expression of genes involved in differentiation, development, proliferation,
and apoptosis. Genes Dev.
15(3):267-85.
47.
Nevins, JR. (2001) The Rb/E2F pathway and cancer. Hum Mol Genet. 10(7):699-703.
48. Nielsen, SJ., Schneider, R., Bauer, U.M., Bannister, A.J., Morrison, A.,
O'Carroll, D., Firestein, R., Cleary, M., Jenuwein, T., Herrera, R.E., Kouzarides,
T. (2001) Rb targets histone H3 methylation and HP1 to promoters. Nature. 412(6846):561-5.
49. Ogawa, H.,
Ishiguro, K., Gaubatz, S., Livingston, D.M., Nakatani, Y. (2002) A complex with
chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science. 296(5570):1132-6.
50. Parakati, R.,
DiMario, J.X. (2005) Dynamic transcriptional regulatory complexes, including
E2F4, p107, p130, and Sp1, control fibroblast growth factor receptor 1 gene
expression during myogenesis. J Biol Chem.
280(22):21284-94.
51. Pohlers, M.,
Truss, M., Frede, U., Scholz, A., Strehle, M., Kuban, R.J., Hoffmann, B.,
Morkel, M., Birchmeier, C., Hagemeier, C. (2005) A role for E2F6 in the
restriction of male-germ-cell-specific gene expression. Curr Biol. 15(11):1051-7.
52. Polager, S.,
Kalma, Y., Berkovich, E., Ginsberg, D. (2002) E2Fs up-regulate expression of
genes involved in DNA replication, DNA repair and mitosis. Oncogene. 21(3):437-46.
53. Powers, J.T.,
Hong, S., Mayhew, C.N., Rogers, P.M., Knudsen, E.S., Johnson, D.G. (2004) E2F1
uses the ATM signaling pathway to induce p53 and Chk2 phosphorylation and
apoptosis. Mol Cancer Res.
2(4):203-14.
54. Rayman, JB.,
Takahashi, Y., Indjeian,V.B., Dannenberg, J.H., Catchpole, S., Watson, R.J., te
Riele, H., Dynlacht, B.D. (2002) E2F mediates cell cycle-dependent
transcriptional repression in vivo by recruitment of an HDAC1/mSin3B
corepressor complex. Genes Dev.
16(8):933-47.
55. Rogoff, H.A.,
Pickering, M.T., Frame, F.M., Debatis, M.E., Sanchez, Y., Jones, S., Kowalik,
T.F. (2004) Apoptosis associated with deregulated E2F activity is dependent on
E2F1 and Atm/Nbs1/Chk2. Mol Cell Biol.
24(7):2968-77.
56. Salomoni, P.,
Pandolfi, P.P. (2002) The role of PML in tumor suppression. Cell. 108(2):165-70.
57. Sanchez, I.,
Dynlacht, B.D. (2005) New insights into cyclins, CDKs, and cell cycle control. Semin Cell Dev Biol. 16(3):311-21.
58. Sanchez-Beato,
M., Sanchez, E., Garcia, J.F., Perez-Rosado, A., Montoya, M.C., Fraga, M.,
Artiga, M.J., Navarrete, M., Abraira, V., Morente, M., Esteller, M., Koseki,
H., Vidal, M., Piris, M.A. (2004) Abnormal PcG protein expression in Hodgkin's
lymphoma. Relation with E2F6 and NFkappaB transcription factors. J Pathol. 204(5):528-37.
59. Semizarov, D.,
Kroeger, P., Fesik, S. (2004) siRNA-mediated gene silencing: a global genome
view. Nucleic Acids Res.
32(13):3836-45.
60. Stanelle, J.,
Stiewe, T., Theseling, C.C., Peter, M., Putzer, B.M. (2002) Gene expression
changes in response to E2F1 activation. Nucleic
Acids Res. 30(8):1859-67.
61. Stevens, C.,
Smith, L., La Thangue, N.B. (2003) Chk2 activates E2F-1 in response to DNA
damage. Nat Cell Biol. 5(5):401-9.
62. Storre, J.,
Elsasser, H.P., Fuchs, M., Ullmann, D., Livingston, D.M., Gaubatz, S. (2002)
Homeotic transformations of the axial skeleton that accompany a targeted
deletion of E2f6. EMBO Rep. 3(7):695-700.
63. Thierry, F.,
Benotmane, M.A., Demeret, C., Mori, M., Teissier, S., Desaintes, C. (2004) A
genomic approach reveals a novel mitotic pathway in papillomavirus
carcinogenesis. Cancer Res.
64(3):895-903.
64.
Trimarchi, JM., Fairchild, B., Wen, J., Lees, J.A. (2001) The E2F6
transcription factor is a component of the mammalian Bmi1-containing polycomb
complex. Proc
Natl Acad Sci U S A.
98(4):1519-24.
65. Valk-Lingbeek,
M.E., Bruggeman, S.W., van Lohuizen, M. (2004) Stem cells and cancer; the
polycomb connection. Cell.
118(4):409-18.
66. Vandel, L.,
Nicolas, E., Vaute, O., Ferreira, R., Ait-Si-Ali, S., Trouche, D. (2001)
Transcriptional repression by the retinoblastoma protein through the
recruitment of a histone methyltransferase. Mol
Cell Biol. 21(19):6484-94.
67. Vernell, R.,
Helin, K., Muller, H. (2003) Identification of target genes of the
p16INK4A-pRB-E2F pathway. J Biol Chem.
278(46):46124-37.
68. Verona, R.,
Moberg, K., Estes, S., Starz, M., Vernon, J.P., Lees, J.A. (1997) E2F activity
is regulated by cell cycle-dependent changes in subcellular localization. Mol Cell Biol. 17(12):7268-82.
69. Wells, J.,
Graveel, C.R., Bartley, S.M., Madore, S.J., Farnham, P.J. (2002) The
identification of E2F1-specific target genes. Proc Natl Acad Sci USA. 99(6):3890-5.
70. Zhang, HS.,
Gavin, M., Dahiya, A., Postigo, A.A., Ma, D., Luo, R.X., Harbour, J.W., Dean,
D.C. (2000) Exit from G1 and S phase of the cell cycle is regulated by
repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell. 101(1):79-89.
71. Zhang, Y.,
Chellappan, S.P. (1995) Cloning and characterization of human DP2, a novel
dimerization partner of E2F. Oncogene.
10(11):2085-93.